Plot all available gates on all flowFrames in a gating set and add layers for the number of events. (In raw format, i.e. *not* re-calculated to volume!)
Usage
plotgates(
gs,
ti = "",
spl = NULL,
fns = NULL,
plotAll = FALSE,
toPdf = TRUE,
x = NULL,
y = NULL,
name.dict = ".",
foN.dict = ".",
type.dict = ".",
foN.plots = "."
)Arguments
- gs
A gating set.
- ti
Character length one, a possible character added to the title of the gate-plot.
- spl
Character length one. The name of the column in the cyTags that should be used to split by before plotting. If left at the default
NULL, no splitting is performed. Possible values for 'spl' are the column names of the cyTags saved in the object of class 'fdmat' as produced bymakefdmat.- fns
Character length one. The filename suffix of the possible pdf.
- plotAll
Logical. If left at the default
FALSE, only the gates where the parameterkeepDatain the gating strategy is set toTRUEare plotted. If set toTRUE, all gates within the gating strategy file will be plotted.- toPdf
Logical. If the plots should be saved in a pdf. Defaults to TRUE
- x
Character length one. The name of channel where data was acquired to be displayed on the x-axis. Only applies if a gating set without applied gate is provided to the argument
gs.- y
Character length one. The name of channel where data was acquired to be displayed on the y-axis. Only applies if a gating set without applied gate is provided to the argument
gs.- name.dict
Character length one. The name of the dictionary. If left at the default '.', the value as defined in the settings file (key 'dD_dict_name') will be used.
- foN.dict
Character length one. The name of the folder where the dictionary resides. If left at the default '.', the value as defined in the settings file (key 'foN_dictionary') will be used.
- type.dict
Character length one. The filetype of the dictionary. Can be one of 'csv' or 'xlsx'. If left at the default '.', the value as defined in the settings file (key 'dD_dict_type') will be used.
- foN.plots
Character length one. The name of the folder where possible PDFs should be saved in. If left at the default '.', the value as defined in the settings file (key 'foN_plots') will be used.
Value
(Invisible) NULL. Is used for its side effect, i.e. to plot gated data resp. to visualize the gating strategy.
Details
Plotting is performed by the function ggcyto.
If a gating set without applied gates is provided to the first argument,
parameters plotAll, spl and toPdf do not apply.
See also
Other Plotting functions:
drawGate(),
plotFlscDist()
Examples
td <- tempdir()
data_source <- "https://github.com/bpollner/data/raw/main/flowdex_examples/flowdex_examples.zip"
check_download_data(td, data_source)
exp_home <- paste0(td, "/flowdex_examples")
old_wd <- getwd()
setwd(exp_home)
#
assign("get_settings_from_flowdex_package_root", TRUE, pos=.GlobalEnv)
# only required to make the examples run automatically
# you should not call 'assign' if you run the examples manually
# the effect of setting 'get_settings_from_flowdex_package_root' to TRUE
# is that the file 'flowdex_settings.R' in 'root' of the installed package
# 'flowdex' will be sourced instead of the one in the user-defined location.
#
gs <- makeAddGatingSet(gateStrat = "gateStrat_2")
#> Reading in fcs files... ok.
#> Producing gating set... Applying fjbiexp transformation... ok.
#> Gating: (6 gates)
#> done!
plotgates(gs, toPdf = FALSE)
#> Coordinate system already present. Adding new coordinate system, which will replace the existing one.
#> .....
#> Coordinate system already present. Adding new coordinate system, which will replace the existing one.
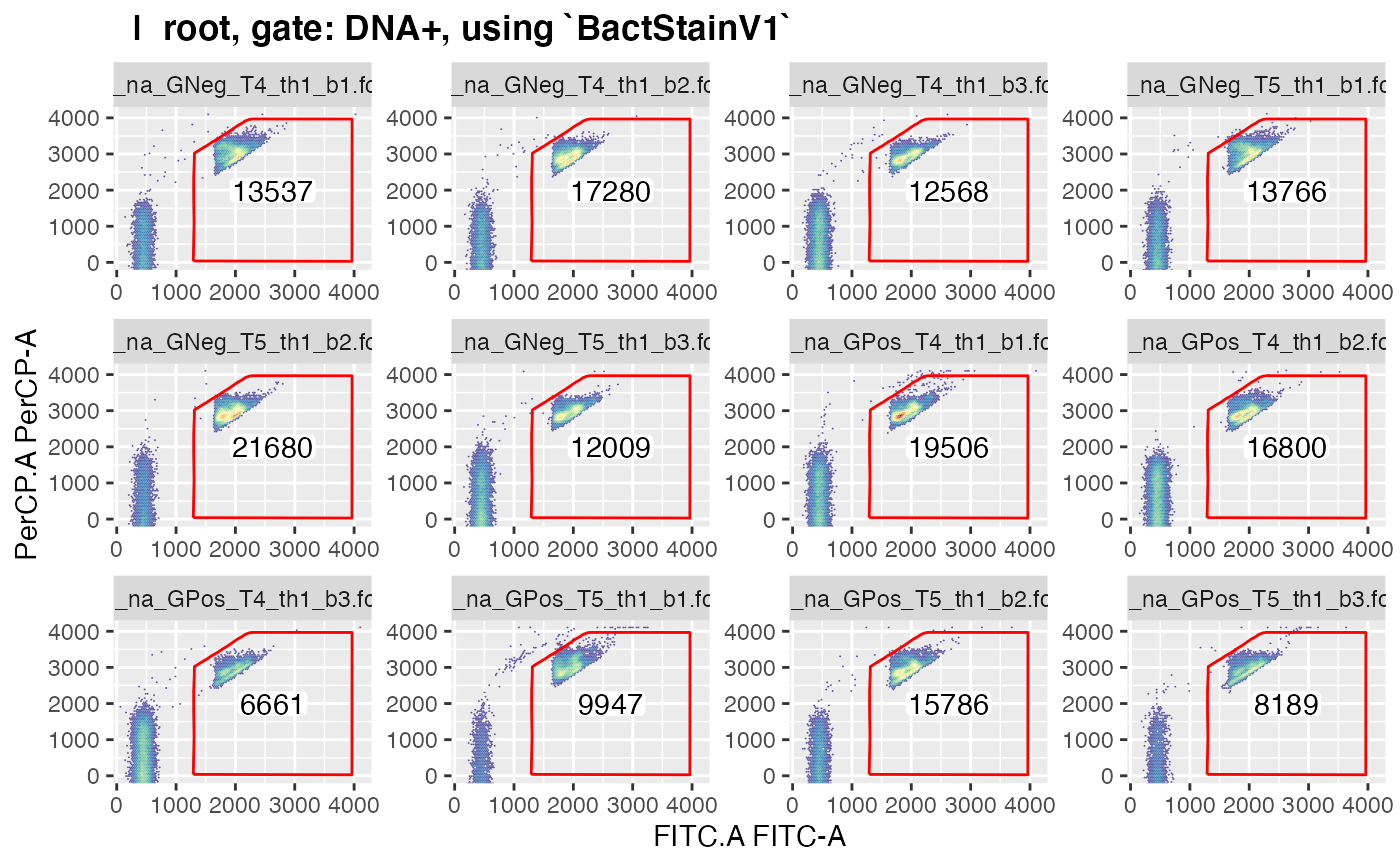
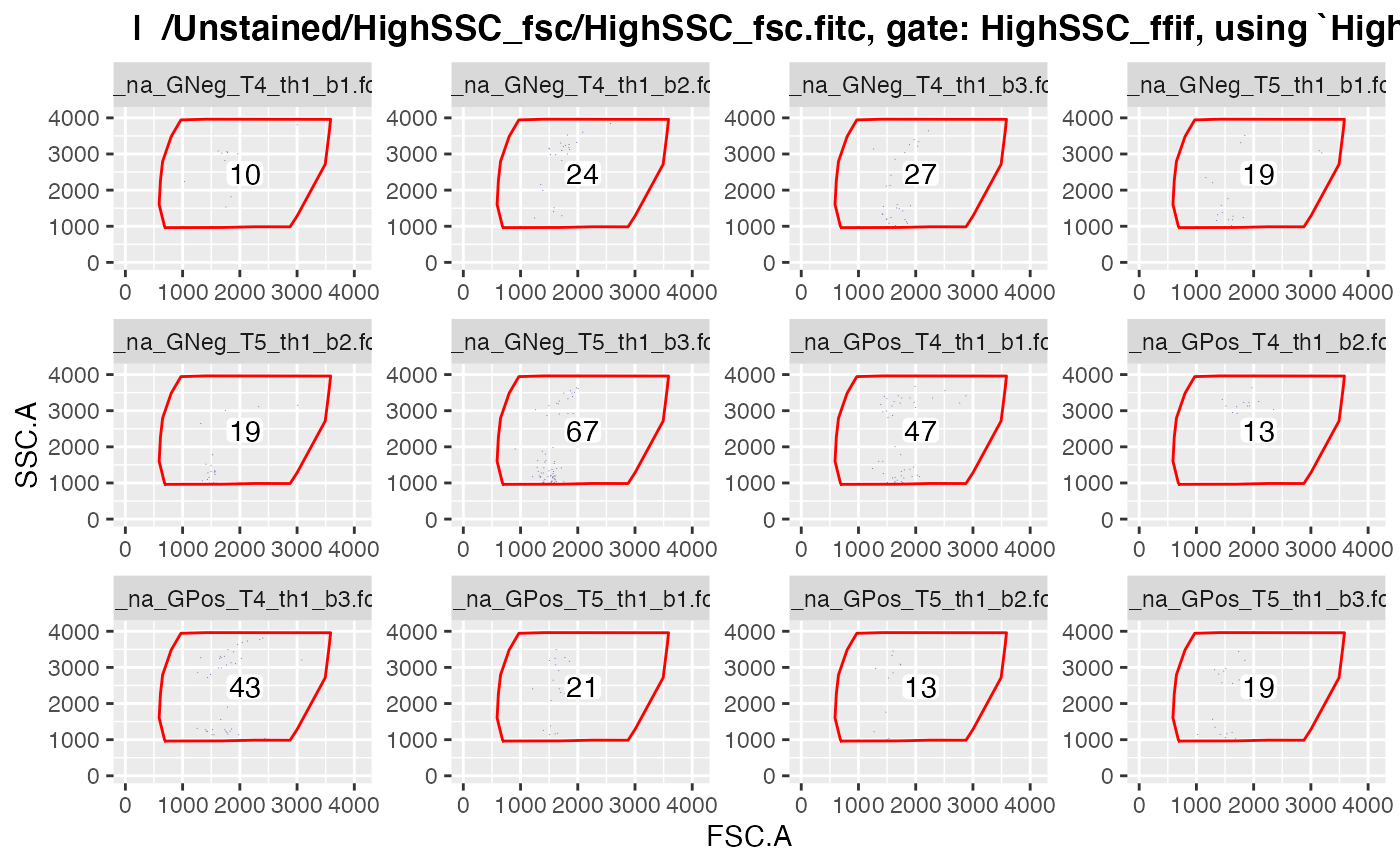 #> .
plotgates(gs, spl = "C_treatment", toPdf = FALSE)
#> Coordinate system already present. Adding new coordinate system, which will replace the existing one.
#> .
#> Coordinate system already present. Adding new coordinate system, which will replace the existing one.
#> .
plotgates(gs, spl = "C_treatment", toPdf = FALSE)
#> Coordinate system already present. Adding new coordinate system, which will replace the existing one.
#> .
#> Coordinate system already present. Adding new coordinate system, which will replace the existing one.
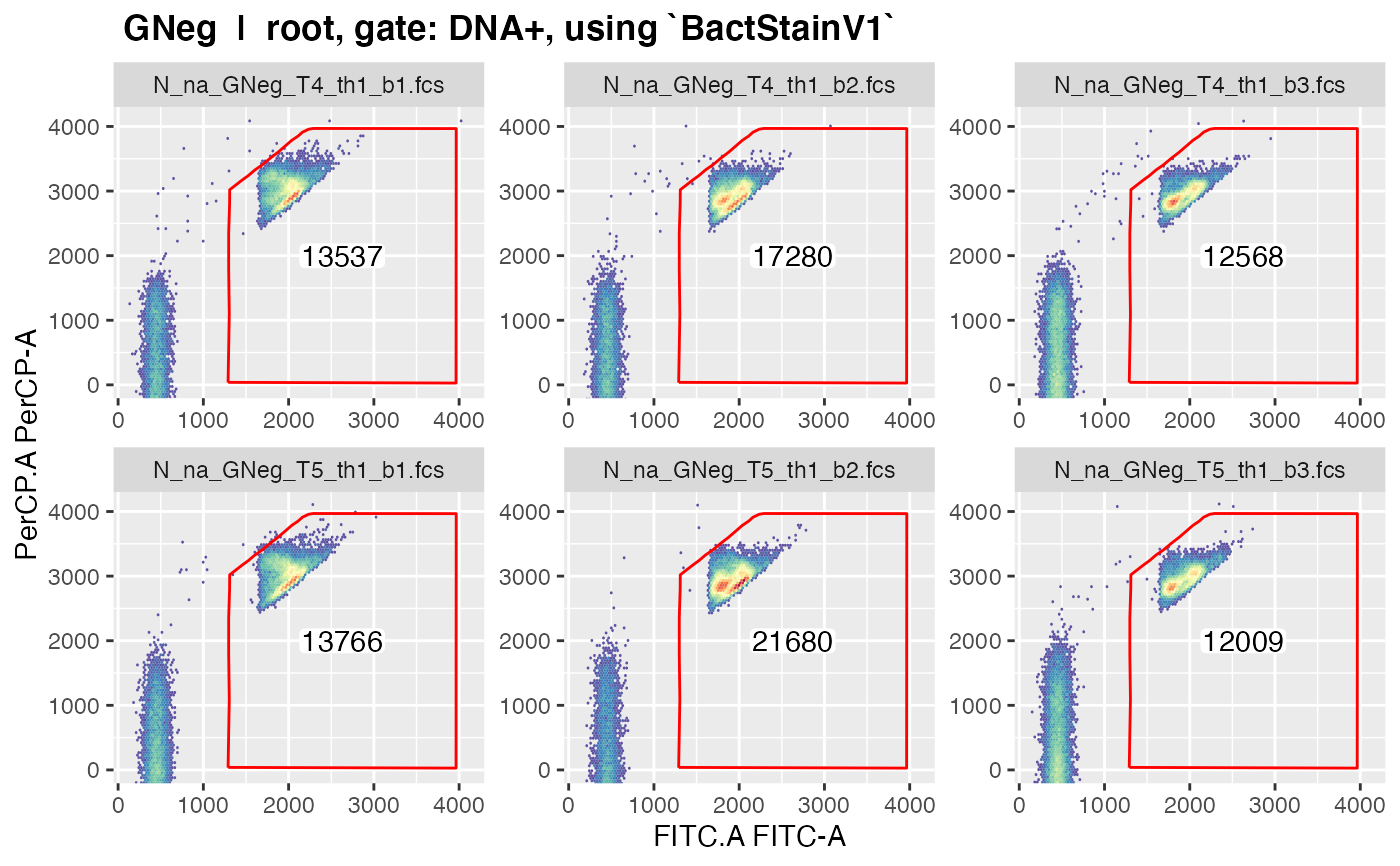 #> ......
#> Coordinate system already present. Adding new coordinate system, which will replace the existing one.
#> ......
#> Coordinate system already present. Adding new coordinate system, which will replace the existing one.
 #> .
#> Coordinate system already present. Adding new coordinate system, which will replace the existing one.
#> .
#> Coordinate system already present. Adding new coordinate system, which will replace the existing one.
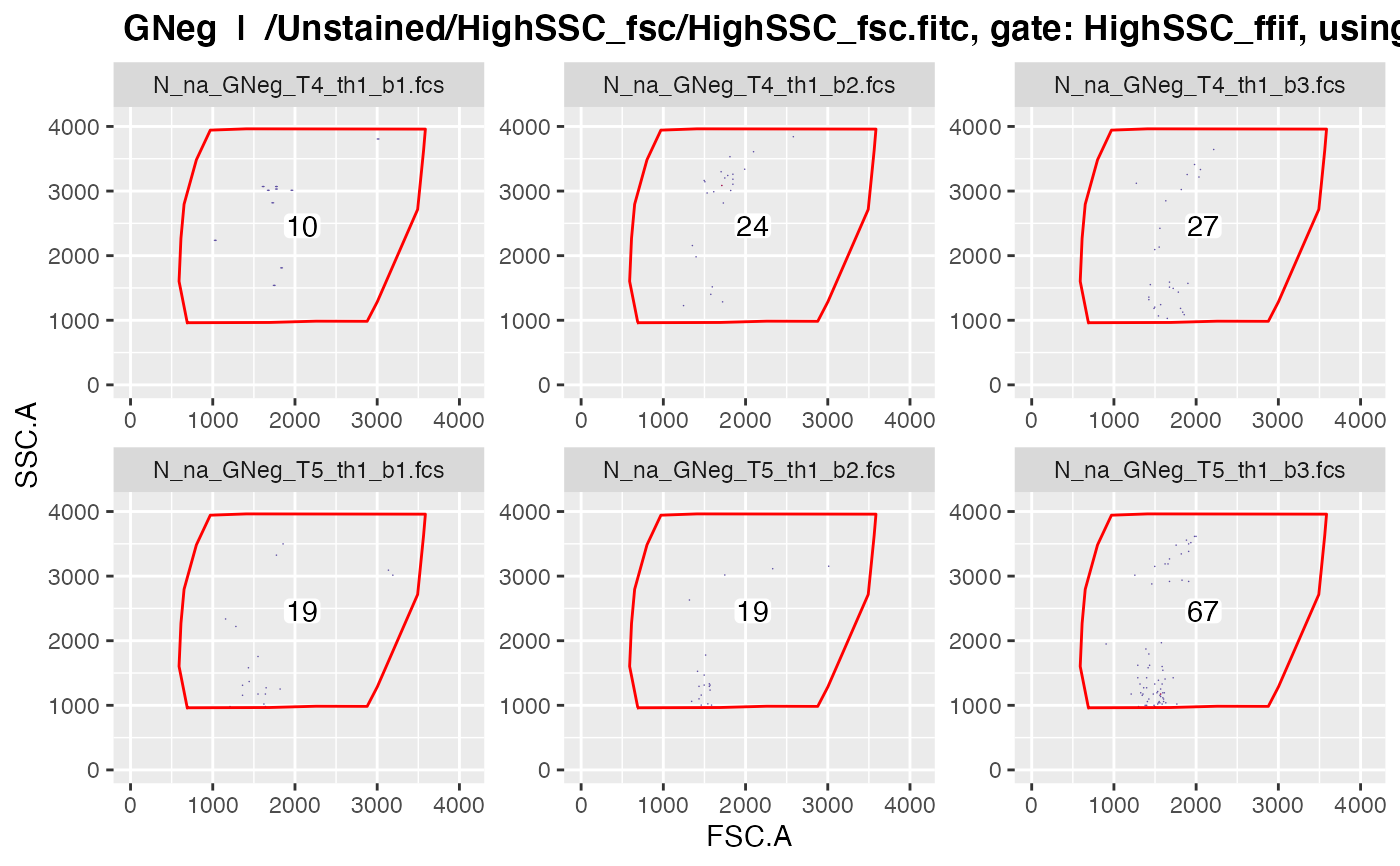
 #> ..
plotgates(gs, spl = "C_treatment", plotAll = TRUE, fns = "_allGates")
#> Plotting gates split by C_treatment...
#> Coordinate system already present. Adding new coordinate system, which will replace the existing one.
#> .
#> Coordinate system already present. Adding new coordinate system, which will replace the existing one.
#> ..
#> Coordinate system already present. Adding new coordinate system, which will replace the existing one.
#> .
#> Coordinate system already present. Adding new coordinate system, which will replace the existing one.
#> ..
#> Coordinate system already present. Adding new coordinate system, which will replace the existing one.
#> .
#> Coordinate system already present. Adding new coordinate system, which will replace the existing one.
#> ..
#> Coordinate system already present. Adding new coordinate system, which will replace the existing one.
#> .
#> Coordinate system already present. Adding new coordinate system, which will replace the existing one.
#> ..
#> Coordinate system already present. Adding new coordinate system, which will replace the existing one.
#> .
#> Coordinate system already present. Adding new coordinate system, which will replace the existing one.
#> ..
#> Coordinate system already present. Adding new coordinate system, which will replace the existing one.
#> .
#> Coordinate system already present. Adding new coordinate system, which will replace the existing one.
#> ..ok.
#
setwd(old_wd)
#> ..
plotgates(gs, spl = "C_treatment", plotAll = TRUE, fns = "_allGates")
#> Plotting gates split by C_treatment...
#> Coordinate system already present. Adding new coordinate system, which will replace the existing one.
#> .
#> Coordinate system already present. Adding new coordinate system, which will replace the existing one.
#> ..
#> Coordinate system already present. Adding new coordinate system, which will replace the existing one.
#> .
#> Coordinate system already present. Adding new coordinate system, which will replace the existing one.
#> ..
#> Coordinate system already present. Adding new coordinate system, which will replace the existing one.
#> .
#> Coordinate system already present. Adding new coordinate system, which will replace the existing one.
#> ..
#> Coordinate system already present. Adding new coordinate system, which will replace the existing one.
#> .
#> Coordinate system already present. Adding new coordinate system, which will replace the existing one.
#> ..
#> Coordinate system already present. Adding new coordinate system, which will replace the existing one.
#> .
#> Coordinate system already present. Adding new coordinate system, which will replace the existing one.
#> ..
#> Coordinate system already present. Adding new coordinate system, which will replace the existing one.
#> .
#> Coordinate system already present. Adding new coordinate system, which will replace the existing one.
#> ..ok.
#
setwd(old_wd)
