Produce a simple x~y dotplot on the specified channels that is
used with locator to define the gate boundaries, similar to defining
a polygon gate in standard FCM GUIs. The resulting data are saved as an
R-object under the name specified in pggId. Press 'esc' when done
drawing the gate.
Usage
drawGate(
gs,
flf = NULL,
gn = "root",
pggId = ".",
channels = ".",
foN.gateStrat = ".",
useLoc = TRUE,
locN = 512,
bnd = ".",
showGate = NULL
)Arguments
- gs
A gating set as produced by
makeGatingSetormakeAddGatingSet.- flf
Character or numeric length one. The identifier of the flowFrame within the gating set where the gate should be drawn on. Optimally, this is a flowFrame, i.e. sample, with a very good representation of the desired population. Possible input values can be determined via 'show'.
- gn
Character length one. The name of a gate further specifying the desired subset of data; defaults to "root".
- pggId
Character length one. The name of the resulting file containing the boundaries of the gate. If left at the default '.', the name as defined in the settings file (key: 'fiN_gate') will be used.
- channels
Character length two. The channels the gate should be defined in. If left at the default '.', the two channels as defined in the settings file (key: 'dV_channelsForPGG') will be used. Available channels can be viewed via 'show' - see examples.
- foN.gateStrat
Character length one. The name of the folder where the file defining the gating strategy and the gate definitions reside. If left at the default '.', the name as defined in the settings file (key: 'foN_gating') will be used.
- useLoc
Logical. If, after plotting, the locator should be used. Defaults to TRUE.
- locN
Numeric length one. How many points to acquire in the locator. Defaults to 512; use "ESC" to abort the locator action.
- bnd
NULL or numeric length four. The boundaries to be marked on the plot, format: (x1, x2, y1, y2). If values are provided, two straight lines on the x-axis and two on the y-axis will be drawn.
- showGate
Character length one. The name of an already existing gate residing in the folder specified by 'foN.gateDefs'. If provided, this gate will be additionally drawn on the dotplot. This can be helpful when e.g. on old, sub-optimal gate should be replaced with a new one: In this case the name of the 'old' would be provided at 'showGate' with the same name specified at 'ppgId'. Thus, the old gate will be replaced with the new one.
Value
A list with the locator coordinates. Mainly called for its side effect, i.e. the locator matrix data saved as an R-object in the folder specified at 'foN.gateStrat'.
Details
The generated R-object is saved automatically and can be used as a gate-definition in the gating strategy. For the lines to be drawn while the locator points are clicked, it is recommended to use this function NOT within R-Studio. The sample names within the gating set can be obtained via 'show' - see examples.
Link
Please refer also to https://bpollner.github.io/flowdex/articles/workflow_1.html for an in-depth description of the workflow how to create the gating strategy.
See also
Other Plotting functions:
plotFlscDist(),
plotgates()
Examples
td <- tempdir()
data_source <- "https://github.com/bpollner/data/raw/main/flowdex_examples/flowdex_examples.zip"
check_download_data(td, data_source)
exp_home <- paste0(td, "/flowdex_examples")
old_wd <- getwd()
setwd(exp_home)
#
assign("get_settings_from_flowdex_package_root", TRUE, pos=.GlobalEnv)
# only required to make the examples run automatically
# you should not call 'assign' if you run the examples manually
# the effect of setting 'get_settings_from_flowdex_package_root' to TRUE
# is that the file 'flowdex_settings.R' in 'root' of the installed package
# 'flowdex' will be sourced instead of the one in the user-defined location.
#
# only required for the automated tests
# this simulates the manual drawing of a gate:
pathToPgg <- paste0(exp_home, "/gating/BactStainV1")
assign("pathToPgg", pathToPgg, pos=.GlobalEnv) # do NOT call this ('assign')
# when running example manually
#####
#####
gs <- makeGatingSet(patt="GPos_T4")
#> Reading in fcs files... ok.
#> Producing gating set... Applying fjbiexp transformation... ok.
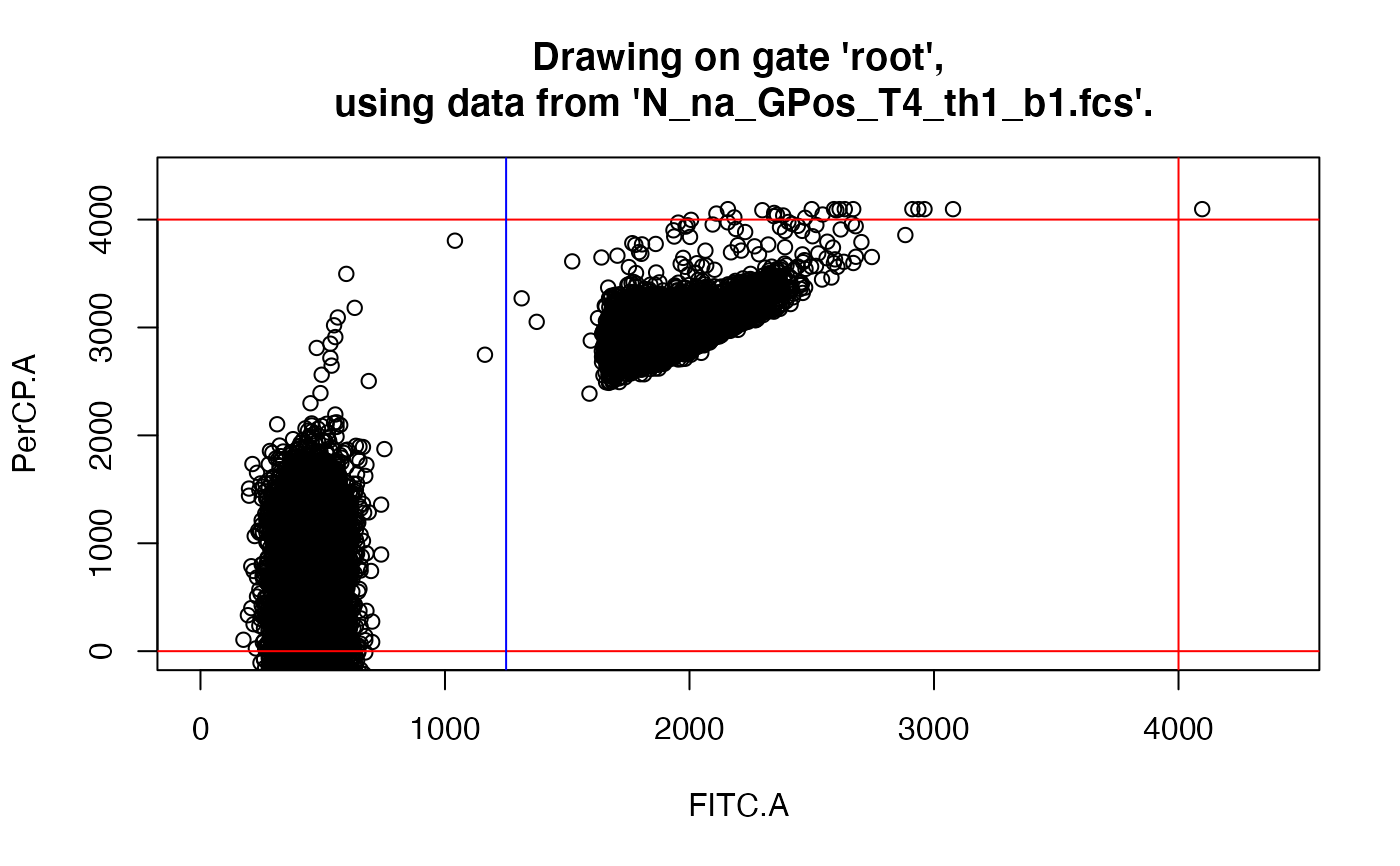
drawGate(gs, 1, useLoc = FALSE) # to just check
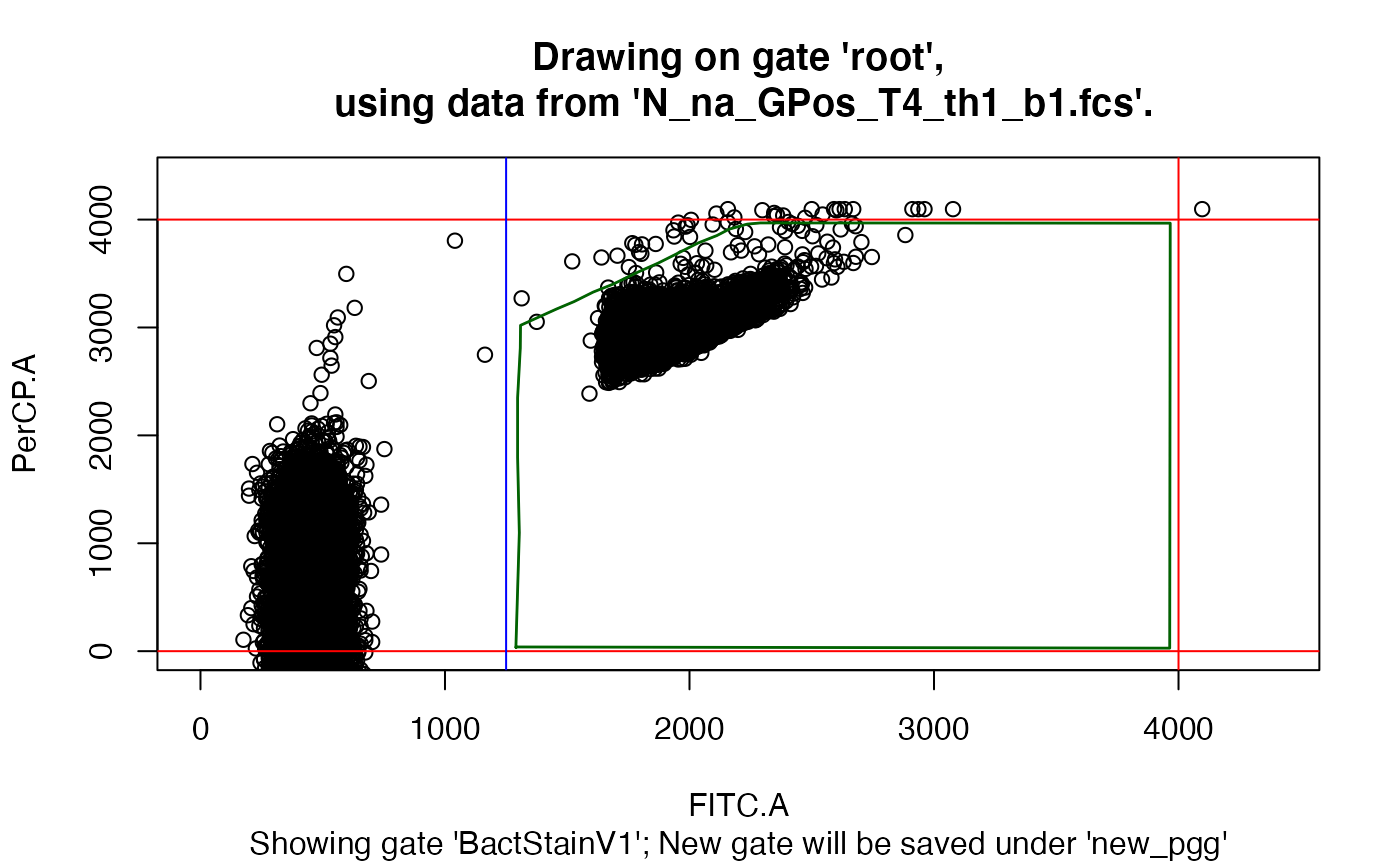
 drawGate(gs, 1, pggId = "new_pgg", showGate = "BactStainV1")
drawGate(gs, 1, pggId = "new_pgg", showGate = "BactStainV1")
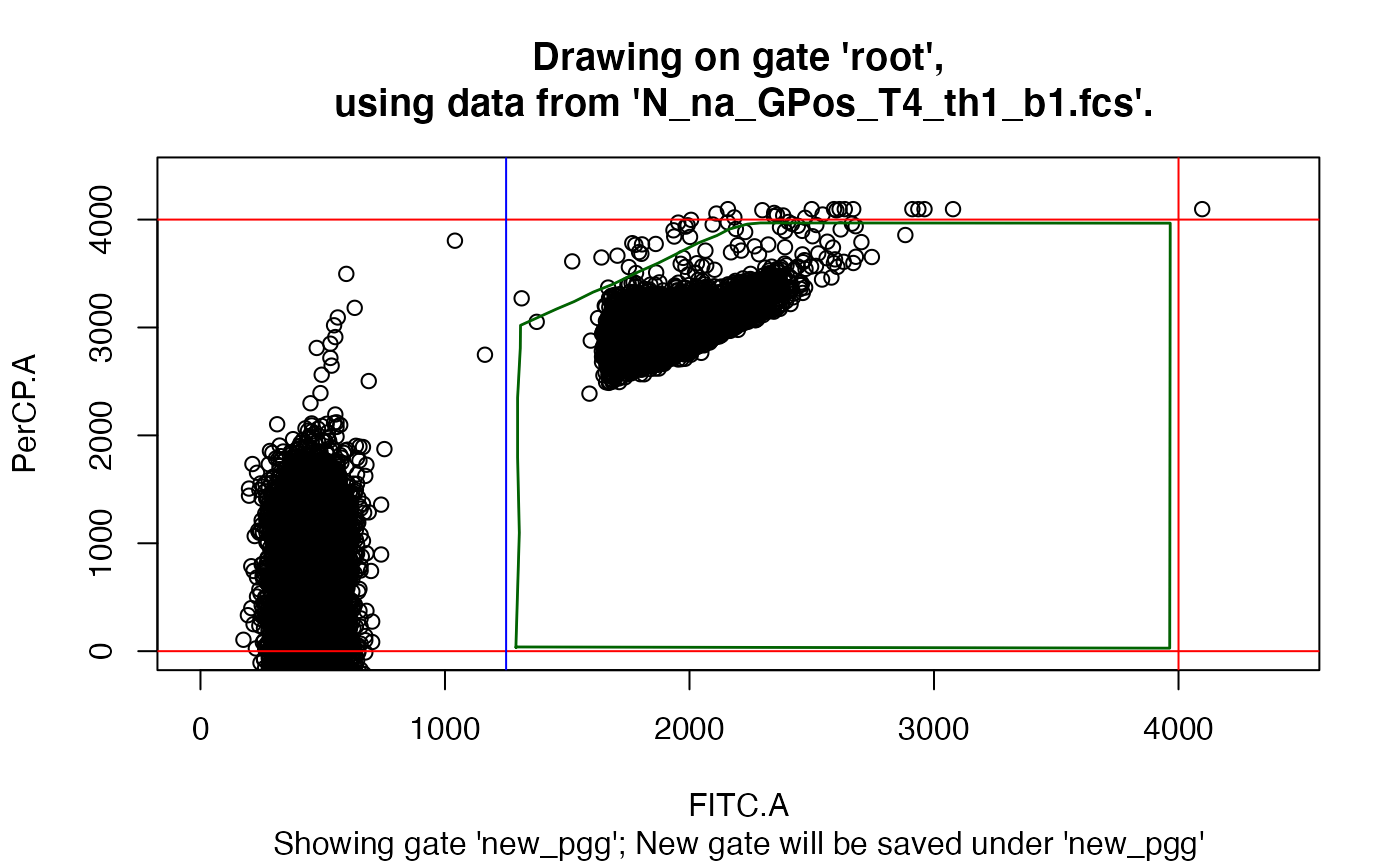
 drawGate(gs, 1, pggId = "new_pgg", showGate = "new_pgg")
drawGate(gs, 1, pggId = "new_pgg", showGate = "new_pgg")
 #
setwd(old_wd)
#
setwd(old_wd)
