Plot the fluorescence distribution contained in the object of class 'fdmat'. For each gate contained in 'fdmat', a graphic will be produced.
Usage
plotFlscDist(
fdmat,
gate = NULL,
ti = "",
spl = NULL,
ylog = FALSE,
ccol = NULL,
clt = NULL,
toPdf = TRUE,
fns = NULL,
foN.plots = ".",
leg = TRUE,
...
)Arguments
- fdmat
An object of class "fdmat" as produced by
makefdmatorflowdexit.- gate
Character or numeric length one. If more than one gate is present in the provided fdmatrix, provide either the name of the gate, or a numeric specifying the position of the gate in the metadata within the fdmatrix to plot data from only that gate. If left at the default
NULLand more than one gate is present in the data, fluorescence distributions from all gates will be plotted in individual plots.- ti
Character length one. Will be used for the title in the plot.
- spl
The column name in the cyTags of the values used for splitting. Defaults to NULL, i.e. no splitting.
- ylog
Logical. If the y-axis (the counts) should be plotted in log scale.
- ccol
An optional color vector for custom coloring. Must have the same length as number of rows in the matrix.
- clt
Numeric vector specifying a sequence of custom line-types.
- toPdf
Logical. If output should be saved in results as PDF. Defaults to TRUE.
- fns
Character length one. The filename suffix, defaults to NULL.
- foN.plots
Character length one. The name of the folder where possible PDFs should be saved in. If left at the default '.', the value as defined in the settings file (key 'foN_plots') will be used.
- leg
Logical, if the legend should be plotted. Defaults to TRUE.
- ...
Additional plotting parameters passed on to 'matplot'
Note
This function is merely intended to give a first overview of the data resp. the fluorescence distribution. Its purpose is not to provide ample and sufficient data visualisation.
Examples
td <- tempdir()
data_source <- "https://github.com/bpollner/data/raw/main/flowdex_examples/flowdex_examples.zip"
check_download_data(td, data_source)
exp_home <- paste0(td, "/flowdex_examples")
old_wd <- getwd()
setwd(exp_home)
#
assign("get_settings_from_flowdex_package_root", TRUE, pos=.GlobalEnv)
# only required to make the examples run automatically
# you should not call 'assign' if you run the examples manually
# the effect of setting 'get_settings_from_flowdex_package_root' to TRUE
# is that the file 'flowdex_settings.R' in 'root' of the installed package
# 'flowdex' will be sourced instead of the one in the user-defined location.
#
fdmat <- flowdexit(patt = "T4")
#> Reading in fcs files... ok.
#> Producing gating set... Applying fjbiexp transformation... ok.
#> Gating: (1 gate)
#> done!
#> DNA+: Extracting binned data on FITC.A (res=220) and recalc. to volume... ok.
#> Exporting data (1 gate) to xlsx...ok.
#> fdmat-object saved.
plotFlscDist(fdmat, toPdf = FALSE)
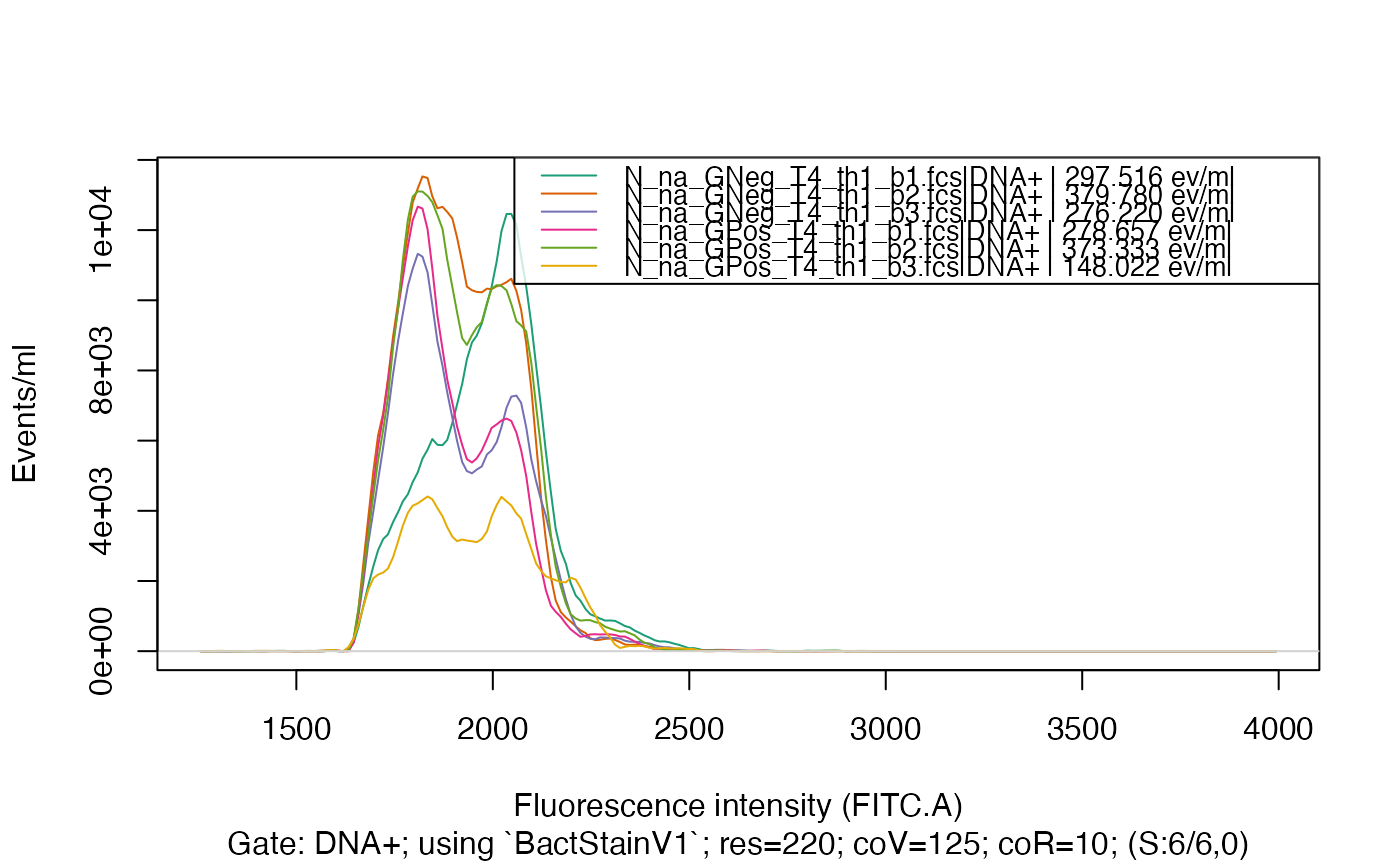 plotFlscDist(fdmat, spl = "C_treatment", toPdf = FALSE)
plotFlscDist(fdmat, spl = "C_treatment", toPdf = FALSE)
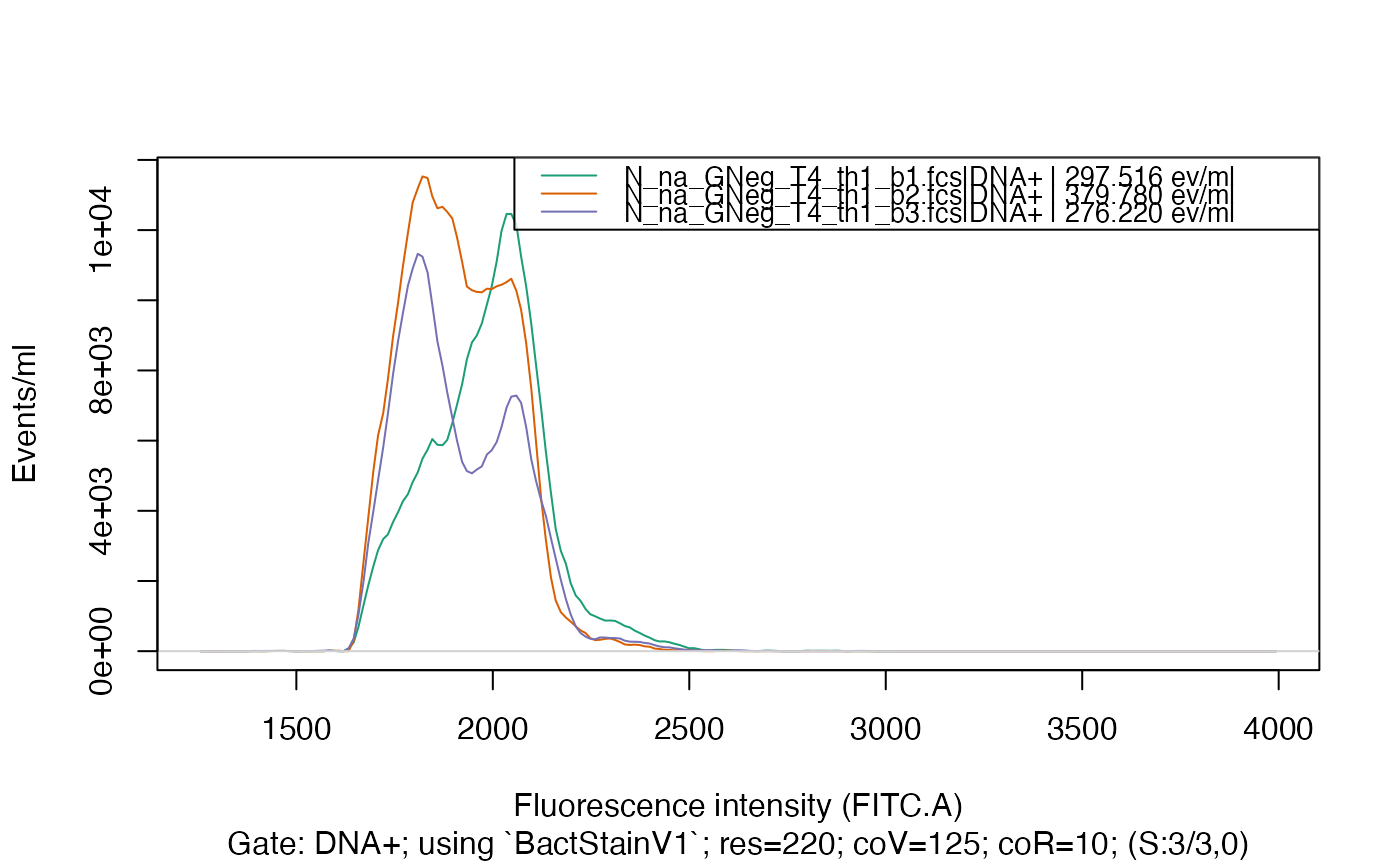
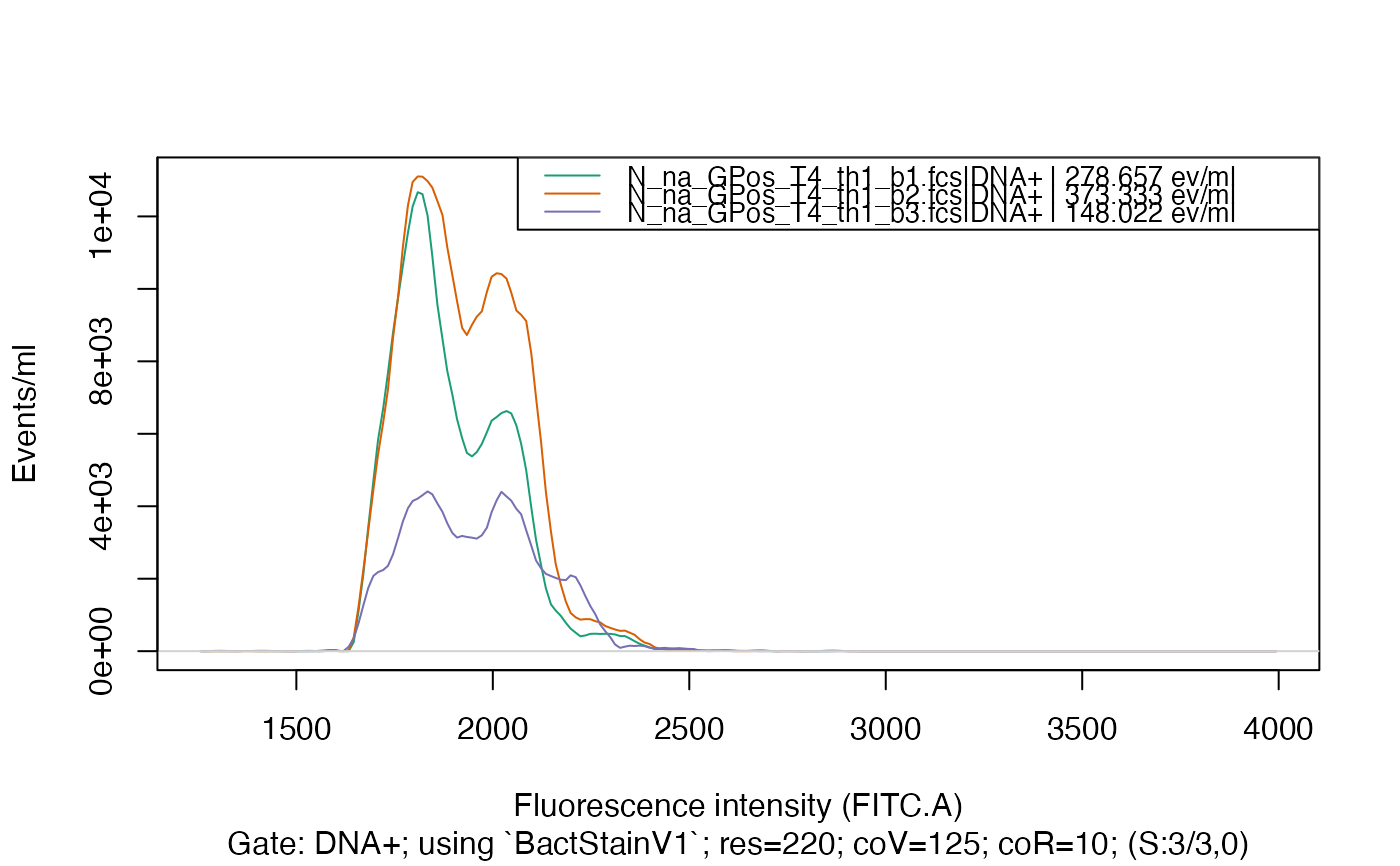 #
setwd(old_wd)
#
setwd(old_wd)
